Abstract
RNA interference (RNAi) is a promising new technology for corn rootworm control. This paper presents the discovery of new gene targets - dvssj1 and dvssj2, in western corn rootworm (WCR). Dvssj1 and dvssj2 are orthologs of the Drosophila genes snakeskin (ssk) and mesh, respectively. These genes encode membrane proteins associated with smooth septate junctions (SSJ) which are required for intestinal barrier function. Based on bioinformatics analysis, dvssj1 appears to be an arthropod-specific gene. Diet based insect feeding assays using double-stranded RNA (dsRNA) targeting dvssj1 and dvssj2 demonstrate targeted mRNA suppression, larval growth inhibition, and mortality. In RNAi treated WCR, injury to the midgut was manifested by “blebbing” of the midgut epithelium into the gut lumen. Ultrastructural examination of midgut epithelial cells revealed apoptosis and regenerative activities. Transgenic plants expressing dsRNA targeting dvssj1 show insecticidal activity and significant plant protection from WCR damage. The data indicate that dvssj1 and dvssj2 are effective gene targets for the control of WCR using RNAi technology, by apparent suppression of production of their respective smooth septate junction membrane proteins located within the intestinal lining, leading to growth inhibition and mortality.
Similar content being viewed by others
Introduction
The western corn rootworm (WCR), Diabrotica virgifera virgifera LeConte (Coleoptera: Chrysomelidae), is one of the most devastating pests in maize that can cause economic losses exceeding $1 billion annually in the U.S.A.1. WCR has traditionally been managed through crop rotation and broad-spectrum soil insecticides2. For over a decade, rootworm management has mainly focused on transgenic corn hybrids expressing Bacillus thuringiensis (Bt) toxins3. Currently, four Bt toxins (Cry3Bb1, mCry3A, eCry3.1Ab and Cry34/35Ab1), are used commercially for the control of WCR and are expressed in corn hybrids either singly or as pyramids4. Recent reports of emerging field insect resistance to both mCry3A and Cry3Bb1 demonstrate the need for effective insect resistance management strategies and discovery of new traits5,6.
RNA interference (RNAi) is a naturally occurring mechanism that regulates gene expression and anti-viral defense in most plants and animals7 and has become an important tool for reverse functional genomics and applications in biomedicine and agriculture8,9. Demonstration of RNA interference following delivery of dsRNA via oral ingestion was first shown in Caenorhabditis elegans10 and has since been documented extensively in insects including WCR11,12,13. Once dsRNA is taken up by cells, dicer RNase III type enzymes bind and digest cytoplasmic dsRNAs into small interfering RNAs (siRNAs) associated with an RNA-induced silencing complex (RISC). The argonaute proteins of RISC cleave the target mRNA strand complementary to their bound siRNA, which determines the specificity of the RNAi activities through precise base-pairing recognition of their complementary target RNAs14. Many genes have been reported to be potential targets in WCR following the provision of dsRNA in diet bioassay13,15,16 and demonstrate that WCR is sensitive to orally delivered dsRNA, providing a new management approach for this important pest17,18.
Pest control via RNA interference has been demonstrated in planta by expressing dsRNA targeted toward the “housekeeping” genes α-tubulin, V-ATPase A13, and C subunit19 or genes involved in cellular pathways such as snf720. V-ATPases are highly conserved multisubunit enzymes that function to acidify intracellular organelles by pumping protons across plasma membranes in exchange for energy21. Snf7 encodes a vacuolar sorting protein involved in intracellular protein trafficking22. Finding new classes of WCR RNAi targets (“modes of action”) is important for effective management of WCR in the future.
The insect midgut plays a critical role in the regulation of important physiological functions such as digestion, metabolism, immune response, electrolyte homoeostasis, osmotic pressure, and circulation23,24. Impairment of one or more of these functions provides a potential basis for new pest management approaches utilizing RNAi. The midgut epithelial cells of most invertebrate species possess specialized cell–cell junctions, known as septate junctions (SJ)25,26, that display a characteristic electron-dense ladder-like structure of 10–20 nm width27. SJs typically form circumferential belts around the apicolateral regions of epithelial cells and control the paracellular pathway26. SJs are subdivided into several morphological types that vary among different animal phyla and different types of SJ have been described in different epithelia within an individual in several phyla25. Molecular and genetic analyses of SJs of invertebrate species have only been performed in Drosophila28,29, where two types of SJ are present: pleated SJ (PSJ) and smooth SJ (SSJ), in ectodermally and endodermally derived epithelia, respectively28. More than 20 PSJ-related proteins have been identified and characterized28. These include transmembrane [e.g. Fasciclin II (FasII), Fasciclin III (FasIII)] and cytoplasmic proteins [e.g. Coracle (Cora), Discs large (Dlg), Lethal (2) giant larvae (Lgl)] localizing at PSJs. In contrast, only two SSJ-specific proteins encoded by Drosophila genes snakeskin (ssk) and mesh have been reported30,31. SSK and MESH form a complex and the two proteins are mutually interdependent for their correct localization31. Several PSJ components, including Dlg, Lgl, Cora and FasIII, have been confirmed to localize to the SSJs. In ssk-and mesh-deficient midguts, Lgl, Cora and FasIII are mislocalized but Dlg is not31. The functions of these PSJ proteins in SSJs remain uncertain since Dlg, Lgl, Cora and FasIII are not required for the SSJ localization of MESH and SSK, and are dispensable for SSJ formation28. The molecular composition of SSJs is different from that of PSJs28. Genetic studies in Drosophila have shown that fluorescent-labeled dextrans (10 kDa) are unable to pass between midgut epithelial cells in wild-type flies but are able to penetrate the paracellular route in mutants defective for smooth septate formation28. The ssk-RNAi and ssk-deletion mutants were lethal at late stage 17 of Drosophila embryo. Ssk and mesh are required for Drosophila development, SSJ formation and midgut paracellular barrier function30,31.
Here we present the discovery of two WCR midgut genes that can potentially serve as effective insecticidal targets using RNA interference technology. Dvssj1 appears to be an arthropod-specific gene that is not found in vertebrates or plants. Insect diet-based assays demonstrated WCR gene target specific mRNA suppression, larval growth inhibition, and mortality. In addition, transgenic maize expressing dsRNA to one of these gene targets (dvssj1) showed a significant reduction in root damage by WCR.
Results
Identification of WCR gene targets
A WCR diet bioassay system for dsRNA-based random screening was developed to identify new and highly active RNAi targets for RNAi-mediated pest control. Double-stranded RNA was produced by in vitro transcription (IVT) and incorporated into WCR diet at a final concentration of 50 ng μl−1 in a 96 well plate format. Insects were scored for mortality and stunting after 7 days and an average primary score was assigned based on 8 observations (replicates) for each dsRNA target. Active target genes (scores ≥ 2) were confirmed and further characterized. Two midgut genes, dvssj1 and dvssj2 (Table 1) were identified among a cohort of 35 WCR RNAi active targets (Supplementary Table 1a).
A set of dsRNA’s targeting dvssj1 and dvssj2, and representing different subfragments of the respective full length sequences were further evaluated in WCR feeding assays to identify fragments with improved efficacy. Fragments with a score ≥2 were selected to determine 50% lethal concentration (LC50) and 50% inhibition concentration (IC50) values (Table 1). Dvssj1 frag1 was the most active dsRNA possessing an LC50 of 0.041 ng μl−1. In contrast, dvssj2 fragments were about 2 to 7-fold less active with a range of LC50 from 0.089 to 0.286 ng μl−1.
Time to 50% lethality (LT50) was measured for dvssj1 and dvssj2 and other active fragments of dvpat3, dvprotb and dvrps10 (see Supplementary Table 2). Dvssj1 and dvssj2 (Table 2) had significantly shorter LT50 than the other active targets. The LT50 (5 ng μl−1) for dvssj1 and dvssj2 was 6.6 and 7.1 days, respectively, compared to LT50 > 8 days for the other active fragments in the assay. Overall, dvssj1 had the shortest LT50 of the RNAi actives tested (Table 2).
SSJ targets are midgut genes
Dvssj1 and dvssj2 were named based on their homology to previously characterized smooth septate junction (SSJ) genes from Drosphilia, ssk30 and mesh31, respectively. DVSSJ1 and DVSSJ2 are 54.9% and 51.3% identical at the amino acid sequence level to SSK (Fig. 1) and MESH (Supplemental Fig. 1), respectively. Both mesh and ssk are required for SSJ formation in the Drosophila midgut30,31. Dvssj1 encodes a 160 amino acid protein with a predicted molecular weight of 17.6 kDa and four predicted membrane-spanning domains (Fig. 1). Dvssj2 has a predicted protein sequence of 1357 amino acids and a MW of 155.9 kDa. The primary structure of DVSSJ2 is similar to MESH in Drosophila31 and contains a single-pass transmembrane (TM) domain, a large extracellular region containing a NIDO (Nidogen-like domain), a TIG (Transcription factor ImmunoGlobin) domain, AMOP32 domain, a VWD (von Willebrand factor type D) domain, and SUSHI repeats (Fig. 2).
The transmembrane domains with hydrophobic residues are indicated by underline predicted by the SOSUI algorithm50. The bold letters indicate the amino acid sequence (TWNLNEEKNPDAEIC) used for monoclonal antibody production. This alignment was derived using CLUSTAL W with default parameters48. * (asterisk) represents identical amino acid residues shared between DVSSJ1 and fly-SSK, : (colon) conservation between two amino acid residues of strongly similar properties and. (period) indicates conservation between two amino acid residues of weakly similar properties.
DVSSJ2 has a single-pass transmembrane (TM) domain, a large extracellular region containing a NIDO (Nidogen-like) domain, aTIG (Transcription factor ImmunoGlobin) domain, an AMOP domain, a VWD (von Willebrand factor type D) domain, and SUSHI repeats predicted by the Pfam protein families database49.
Target expression and suppression by dsRNA feeding
Ssk and mesh in Drosophila are specifically expressed in endodermally derived epithelia, including the midgut, gastric caeca, the outer epithelial layer of the proventriculus, and the Malpighian tubules30,31. Proteins of both dvssj1 and dvssj2 were detected in midgut homogenates extracted from 3rd instar WCR with expected MWs (arrows) of 17.6 and 155.9 kDa, respectively (Fig. 3a and Supplementary Fig. 2).
(a) Western blot detection of DVSSJ1 and DVSSJ2 from 3rd instar WCR dissected gut tissues. Loaded samples represent the equivalent of 1 WCR midgut. Detection of DVSSJ1 is by anti-DVSSJ1 monoclonal peptide antibody (Peptides sequence: TWNLNEEKNPDAEI 41-53 a.a.). DVSSJ2 was detected by Hybridoma Supernatant from peptide antibody production. (Peptide Sequence: MTSDTAPPDTDQRG 108-121 a.a.). The DVSSJ1 and DVSSJ2 detectable protein sizes are compared by Precision Plus Protein Western Standard (Std, Bio-Rad) ranging from 10–250 kDa (Supplementary Fig. 2). (b) Relative gene expression of dvssj1 over time by dsRNA treatment. qRT-PCR was used to examine gene expression of dvssj1. The expression represented relative expression for time points 12 and 48 hrs for treatments of double stranded RNA for dvssj1 and gus. H20 was used as a control treatment. Relative expression analysis was based on dvssj1 expression, after being normalized by reference gene dvrps10 expression, and then compared to dvssj1 expression in H20 control at each time point. Each value was shown as values ± S.E.M. of individual insects. Letter differences represent treatments that are significantly different from each other (P-value < 0.05) determined by one-way analysis of variance (ANOVA) followed by Tukey’s post-test.
Suppression of dvssj1 mRNA accumulation was demonstrated using both quantitative RT-PCR and in situ hybrization (ISH) methods. Expression of dvssj1 mRNA was quantified from insects exposed to 0.5 ng μl−1 of diet incorporated dvssj1 dsRNA and collected after 12 and 48 h of feeding. Dvssj1 mRNA accumulation was significantly (P < 0.05) reduced following dvssj1 dsRNA treatment but not with a control dsRNA (gus) or water control (Fig. 3b). Dvssj1 expression was lowest 48 h post treatment. Localization of dvssj1 mRNA molecules in 3rd instar WCR was demonstrated using RNAscope ISH. Dvssj1 mRNA molecules were predominantly present in the cells of the midgut epithelium (Fig. 4) but were also detected in the oenocyte cells33. At 48 h post-treatment of dvssj1 dsRNA, midgut epithelium cells showed a loss of dvssj1 mRNAs. The distal oenocyte cells also showed a nearly complete loss of dvssj1 mRNAs.
Representative midgut sections (Supplementary Fig. 3) were from WCR 3rd instars treated with H2O (top panel), gus dsRNA (middle panel) and dvssj1 frag1 (bottom panel) at 50 ng μl−1 for 48-h. All treatments were hybridized with the dvssj1 probe and an RNAscope® negative control probe (Bacillus subtilis dihydrodipicolinate reductase (dapB) gene). Expression of dvssj1 mRNA is observed in midgut epithelium cells (ep) and oenocyte cells (o) of H2O and control gus treatment. Knockdown of dvssj1 mRNA in midgut epithelium cells (ep) and oenocyte cells (o) is observed in larvae treated with dvssj1 dsRNA (bottom panel). No clear presence of dvssj1 mRNA in fat body cells (f) and dvssj1 dsRNA in midgut lumen was observed (arrow). Images were captured at 40× magnification with 100 μm scale bars.
Ultrastructure observations
After 72 h of feeding, dvssj1 dsRNA treatment (100 ng μl−1) resulted in an overall decrease in neonate length compared to untreated controls (Supplementary Fig. 4a,b); in some cases, the gut lumen volume was greatly reduced by the presence of apparent blebs from intestinal epithelial cells. Unique ultrastructural features very rarely or not observed in corresponding untreated controls (cf. Supplementary Fig. 5a–e) indicative of midgut epithelial cell injury were found in treated neonates at 72 h. These features included apparent manifestations of apoptosis and accelerated regenerative activities such as unusual stem cell morphology, reduction in basal extracellular labyrinth, and appearance of numerous vesicles in different regions of enterocyte cytoplasm (Fig. 5a–d).
(a) Anterior midgut epithelium exhibiting apoptotic enterocytes (En) that mostly lack basal extracellular labyrinth, but contain highly vesiculated cytoplasm. Numerous stem cells are present, typically containing electron dense cytoplasmic inclusions. A “dark body” (Db) consisting of nascent microvilli is taken as evidence of an active regenerative process. Mv, microvilli (b) Higher magnification of a central area taken from panel a to better illustrate stem cell detail. Nu, nucleus (c,d) Epithelium from anterior/middle and middle midgut regions. Note apoptotic enterocytes with highly vesiculated cytoplasm either lacking or with reduced, partially vesiculated (Vd) basal extracellular labyrinth. Regenerating cells (*) are interspersed among sloughing cells and stem cells (Sc). Bm, basal membrane; Non-osmicated specimens.
Dvssj1 provides root protection against WCR
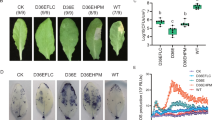
Transgenic maize lines expressing dvssj1 dsRNA were generated through transformation of a Pioneer inbred line, PHR03. Five transgenic events expressing dvssj1 dsRNA were selected for greenhouse assay at the T1 generation (14–15 plants per event) and infested with 1000 WCR eggs at the V6 (six-leaf) stage. Plants were scored for WCR feeding damage34 three weeks after infestation. The average node injury scores for the transgenic events were 0.12–0.61, which was a significant (P < 0.0001) reduction from the corresponding score of 1.5 for the negative control isoline (Fig. 6 and Supplementary Table 4).
(a) Map of the dvssj1 expression cassette. (b) Five dvssj1 transgenic lines and one transgenic negative isoline (NULL) were selected for T1 greenhouse assay. Fifteen or fourteen plants per dvssj1 dsRNA line and NULL plants were assayed for WCR feeding damage34. The node injury score (mean ± SD) was significantly different (p-value < 0.0001) between the NULL and all transgenic lines.
Total RNA was extracted from root tissues for northern blot analysis to examine RNA expression in the T1 transgenic plants. Two species of RNA were detected by the dsRNA northern blot (Fig. 7) - a dominant band migrating at approximately 232 nucleotides (nt) and a less intense band migrating higher than 232 nt. The 232 nt dominant band likely represents dsRNA (Supplementary Fig. 6b). DsRNA transcripts have previously been reported in maize13,19. Dvssj1 dsRNA derived small RNAs (21 to 24-nt RNAs) were identified on a siRNA northern blot (Fig. 7). The prevalent species of siRNA appeared to be 21 nt fragments, consistent with previous findings in transgenic maize containing RNAi constructs13,19. Expression of dvssj1 RNAs correlate to the copy number of the transgene and has an inverse relationship with nodal injury score (Fig. 7 and Supplementary Table 4).
Northern blots of five dvssj1 containing events and one transgenic negative isoline (NULL). Zmactin (Accession #: EU952376; top panel) was included as a reference gene for northern analysis. Double-stranded dvssj1 frag1 (210 bp; 100, 50 and 5 pg) RNAs were loaded as a positive control of dsRNA (middle panel); 29nt dvssj1 oligo (100, 50 and 5 pg) was used as a positive control for the siRNA northern blot (bottom panel).
Discussion
In order to screen for new RNAi targets in WCR we developed a 7-day diet based assay. Two key factors were employed for an efficient and reliable diet assay: (1) DsRNAs were incorporated into standard WCR artificial diet containing food coloring to monitor feeding, and (2) only healthy larvae were selected for the screening. Using this assay, thirty-five RNAi active targets were identified (Supplementary Table 1a). These targets included members of several gene families that have been previously reported13,17, such as ribosomal proteins, proteasome subunits, transcription and translation initiation elongation factors35. Other notable RNAi gene targets included members of the small GTPase superfamily36, heat shock proteins, and actin genes13,15. Two unique genes, dvssj137 and dvssj238, initially identified as effective RNAi targets were subsequently found to be orthologs of Drosophila ssk30 and mesh31 proteins, respectively (Table 1, Figs 1 and 2). The first SSJ gene was reported in Bombyx mori (silkworm) using monoclonal antibodies that specifically recognized the apical region of the lateral membrane of midgut epithelial cells. The monoclonal antibodies were subsequently used to immunoprecipitate proteins from a midgut membrane fraction followed by protein sequence determination using mass spectrometry30. The Drosophila ortholog was identified by homology search and named Snakeskin (ssk). SSK has four membrane-spanning domains predicted in its primary protein sequence. Another SSJ gene, mesh, was also identified using a similar approach31. Proteins of both genes are important to the formation of the SSJ in the Drosophila midgut28.
In Drosophila, SSK and MESH colocalize to SSJ and are specifically expressed in endodermally derived epithelia, including the midgut and gastric caeca30,31. Western analyses confirmed that both DVSSJ1 and DVSSJ2 were present in WCR midgut derived tissues (Fig. 3a). Several higher MW bands were visible in the DVSSJ1 western blot which were likely non-specific signals, protein complexes with DVSSJ231 or complexes with other unidentified proteins. SSK expression appears at stage 12 of Drosophila embryos in midgut rudiments as a protein band of ~15 kDa and its expression is sustained until the adult stage throughout the midgut and Malpighian tubules30. The lower MW band in the DVSSJ2 western blot may represent truncated version or a form of split variant. There are five different mesh variants in Flybase39, which translated into three isoforms with different C-terminal cytoplasmic regions. MESH was detected by western analysis as a main ~90 kDa band and a minor ~200 kDa band31. Compromised mesh expression causes defects in the organization of SSJs, resulting in the mis-localization of other SSJ proteins, and the loss of barrier function of the midgut. Ectopic expression of MESH in cultured cells induces cell-cell adhesion. Drosophila SSK and MESH form a complex together and these proteins are mutually interdependent for their correct localization in SSJ formation31.
Quantitative RT-PCR analyses confirmed that ingestion of dvssj1 dsRNA resulted in suppression of dvssj1 mRNA (Fig. 3b). Dvssj1 mRNA expression patterns and dvssj1 mRNA knockdown were also demonstrated in 3rd instar WCR using RNAscope ISH (Fig. 4). Dvssj1 only expressed in midgut epithelium cells and oenocyte cells, and expression patterns varied slightly between different regions of the midgut (Supplementary Fig. 3). The gene expression of dvssj1 mRNA in midgut epithelium cells of WCR corroborates the functional role of dvssj1, analogous to Drosophila ssk30. Dvssj1 mRNA were also detected in oenocyte cells, which are cells responsible for lipid processing and detoxification40.
The physical integrity of the SSJ is important for controlling the paracellular pathway between epithelial cells, which effectively separates the gut lumen, where digestion occurs, from the interstitial space, where metabolites and electrolytes are tightly regulated25. The SSJ is composed of a group of proteins physically connecting adjacent cells and contribute to the specialization between epithelial cell apical and basolateral membranes28. Although the molecular architecture of the WCR SSJ has not been fully characterized, DVSS1 protein is clearly an ortholog of the integral membrane protein (SSK) in Drosophila30. Mutant flies lacking ssk do not survive early larval development28. Flies with reduced ssk expression exhibit deformed midgut epithelial cells and uncontrolled leakage of a tracer dye from the gut into the hemocoel30. Similarly, suppression of mesh has the same effect on fly midgut epithelium31. The extracellular domains of MESH are found in cell adhesion proteins that are involved in cell-cell and cell-matrix adhesion28,31. The toxic effect to WCR resulting from oral exposure to dvssj1 dsRNA in diet or expressed in planta can be attributed to suppression of dvssj1 mRNA leading to reduction in DVSSJ1 expression/accumulation, loss of the midgut epithelium diffusional barrier, and cellular deformities due to improper intercellular contacts. Future studies may include quantitative analyses of dvssj1 mRNA and protein from insects exposed to different doses of dsRNA to help understand dvssj1 RNAi effects and target protein stability or turnover rate.
Cytological observations of nearly whole neonates in section (Supplmentary Fig. 4) showed a significant difference in the overall size between treated and untreated individuals, as well as an apparent occlusion of the gut lumen, and numerous examples of enterocyte blebbing into the gut lumen, after dvssj1 dsRNA consumption. Extensive comparison of dsRNA-treated neonate sections to sections prepared from untreated controls was important for distinguishing between normal cell regeneration and molting, and potential effects of dvssj1 dsRNA treatment on neonate mid-gut epitheial cell ultrastructure. For example, dark bodies that contained what appeared to be nascent microvilli41 were very often observed at 72 h in dsRNA-treated neonate samples (Fig. 5), but rarely in the controls (Supplementary Fig. 5). In gut areas, where much blebbing of enterocyte cytoplasm into the gut lumen could be observed, basal extracellular labyrinth (Bl) was sometimes not evident. Such regions also exhibited enlarged and differentiating stem cells which we interpreted as evidence of active molting or possible stress response42. These regions, which also bore additional subcellular markers such as dark bodies as mentioned above, and highly vesiculated cytoplasm, were especially prevalent in dvssj1 dsRNA-treated larval gut, and only rarely observed in controls. These observations are consistent with the notion that suppression of dvssj1 expression and its protein accumulation are the cause of WCR growth inhibition and mortality.
SSK and MESH are improtant to the formation of the SSJ in the midgut of Drosophila30,31. Ssk orthologs have been identified in other arthropods, but not in vertebrates30, suggesting that SSJs composed of MESH and SSK are arthropod-specific cell–cell junctions. However, MESH homologs are present in other metazoans, including C. elegans, sea urchins and mammals31. A sequence search of public and internal databases suggests that dvssj1 orthologs are only found in arthropods and not in vertebrate species or maize (Supplementary Fig. 6 and Supplementary Table 5). This makes dvssj1 a good target for applying RNAi rootworm control in transgenic plants.
Dvssj1 dsRNA targets the expression of a protein important for the formation of SSJ between epithelial cells lining the midgut. Ingested Dvssj1 dsRNA has a relatively fast biological effect on WCR as indicated by its short LT50 (Table 2) which may be a consequence of direct exposure of midgut epithelial cells to dsRNA. Consequently, the biological effect of dvssj1 RNAi may have no dependency on systemic movement of the silencing signal11. During WCR larval development, the midgut epithelial surface area grows by the continuous increase of the number of cells43. Maintenance of midgut epithelial characteristics during this period requires tightly regulated SSJ to support vital structure and barrier functions. Disruption of SSJ by the down regulation of dvssj1, makes this a well-suited gene target for RNAi silencing and an alternative “mode of RNAi action” for the control of corn rootworm. Under greenhouse conditions, mean node injury scores for the transgenic dvssj1 events ranged from 0.12–0.61, which was a significant (P < 0.0001) reduction from the corresponding score of 1.50 for the negative control isoline (Fig. 6 and Supplementary Table 4). Dun et al.44 and Tinsley et al.45 estimated that under field conditions, one node of root injury was on average associated with a corn yield loss of approximately 15–18%. Further studies are needed to confirm the efficacy46 of dvssj1 events under field conditions47.
Conclusion
The discovery of dvssj1 and dvssj2 genes in WCR provides new potential gene targets or “modes of action” at the gene level for the control of this important pest using RNA interference technology. Double-stranded RNA targeting dvssj1 expressed in transgenic maize plants can effectively down regulate the expression of the dvssj1 gene in WCR larvae, leading to larval growth inhibition and mortality.
Methods
WCR cDNA library and identification of RNAi active clones
The cDNA library construction kit from Clontech (Mountain View, CA) was used to make WCR cDNA libraries. Total RNA was extracted from WCR neonates or 2nd–3rd instars and cDNAs were cloned into the Sfi I site of the pDNR-LIB library vector according to the manufacturers’ instructions. Expressed sequence tag (EST) sequencing was performed using Applied Biosystems capillary sequencers. RNAi target screening includes primary screening (8 replicates for each target) and confirmation (8 replicates per target) round, which was conducted on subset of primary active targets based on primary score (activity) and target novelty. Dose response assays were used for further characterizing insecticidal activities in diet. After an active target (cDNA clone) was identified via tBLASTx against both Tribolium and/or Drosophila database, full length cDNA was sequenced using standard Sanger sequencing methods or transcript was identified from the WCR transcriptome analysis (Supplementary Table 1a). Sequence alignment was derived using CLUSTAL W with default parameters48. Protein domains were predicted by the Pfam protein families database49. The transmembrane domains with hydrophobic residues were predicted by the SOSUI algorithm50.
Double stranded RNA production by in vitro transcription
To screen WCR active targets, 400 to 800 base pair regions of randomly selected non- redundant cDNA clones were amplified using Taq DNA polymerase with a pair of gene specific primers (Supplementary Tables 1b and 2). The gene-specific primers also contained T7 RNA polymerase sites (5′d[TAATACGACTCACTATAGGG]3′) at the 5′ end of each primer. PCR product served as the template for dsRNA synthesis by in vitro transcription (IVT) using a MEGAscript kit (Life Technologies, Carlsbad, CA). DsRNAs were examined by 48 well E-gel electrophoresis (Life Technologies) to ensure dsRNA integrity and quantified using Phoretix 1D (Cleave Scientific).
WCR bioassays
Diet-based bioassays for primary screening
WCR diet was prepared according to the manufacturer’s guideline for Diabrotica diet (Frontier, Newark, DE) with modifications51. DsRNA samples were incorporated into diet at 50 ng μl−1 final concentration in a 96 well microtiter plate format. In each well of the plate, a mixture of 5 μl of dsRNA (300 ng μl−1) and 25 μl of WCR diet were added to each well of the plate and shaken on an orbital shaker for 1 minute until the diet solidified. Eight replicates (wells) were used for each RNA sample. Preconditioned 1st instar WCR (neonates were placed on diet for 24 h prior to transfer to the test plate) were added to the 96 well plates; 2 insects per well. After 7 days of incubation, larvae were scored for growth inhibition and mortality using the following scale: 0 = No effect, larvae are equal to control plate larval growth (2nd instars), 1 = Slight larval stunting, larvae are slightly smaller (i.e ~25% reduction) in length and width, 2 = Severe larval stunting, larvae are 1st instars (approx. the size of the infested neonates or >60% reduction in size of healthy insects), 3 = Dead (100% Mortality). The primary and confirmation scores were based on an average score across all eight replicates.
LC50 and IC50 determination
Double-stranded RNAs were incorporated in diet as described for primary screening. For each sample, ten doses (100, 31.6, 10, 3.16, 1, 0.316, 0.10, 0.032, 0.010 and 0.0032 ng μl−1) were evaluated for a total of 32 observations per dose or water control. Four plates were employed with 8 wells on each plate for each concentration. Two one-day old larvae were transferred into each well. Plates were incubated at 27 °C and 65% RH. Seven days after exposure larvae were scored for growth inhibition (severely stunted larvae with >60% reduction in size) and mortality. Data were analyzed using PROC Probit analysis52 in SAS to determine the 50% lethal concentration (LC50). The total numbers of dead and severely stunted larvae were used for analysis of the 50% inhibition concentration (IC50).
Lethal Time (LT50) determinations
LT50 assays were performed on five WCR dsRNA target genes, with two different assays for each sample. In the first assay, the same method as was described for the LC50/IC50 determinations was used. A single one-day old larva was transferred into each well of a 48 well plate containing 400 μl of diet with dsRNA sample at 5 ng μl−1. In the second assay, a single one-day old larva pretreated on diet containing 50 ng μl−1 of dsRNA was transferred into each well of a 48 well plate containing 400 μl of diet and dsRNA at the same dose for each sample. The plates were scored daily for mortality 1–12 days after infestation. The Weibull distribution for Survival analysis in SAS (Version 9.4) was used to describe the time to mortality curve. Each insect was treated as an individual data point for the LT50 output based on a Weibull distribution. LT50’s were considered significantly different if 95% confidence intervals (CI) (P < 0.05) were non-overlapping.
Plant expression vectors and transformation
Standard DNA and RNA techniques as described by Sambrook and Russell53 were used for vector construction and expression analyses. To demonstrate rootworm efficacy in planta, a fragment of the dvssj1 gene was assembled into a suppression cassette designed to express dsRNA targeting a section of the dvssj1 gene. The silencing cassette consisted of the maize ubiquitin promoter, maize ubiquitin intron 154, two 210 base pair stretches of dvssj1 and an intervening truncated maize ADH intron1 designed to support assembly into a dsRNA hairpin (Fig. 6a and Supplementary Fig. 6b), and the PIN II terminator. The dvssj1 construct was transformed via Agrobacterium tumefaciens into a commercial maize elite-inbred line, PHR0355. T0 maize transformants were screened by qPCR analyses56 and transferred to soil and backcrossed with a PHR03 inbred line to generate T1 progeny.
Greenhouse WCR feeding assays
T1 plants containing one or more copies of the dvssj1 silencing cassette were evaluated using a greenhouse assay to assess rootworm feeding damage. T1 seeds were planted in 32-cell flats containing Fafard Superfine potting mix. Fifteen or fourteen PCR positive plants (Supplementary Table 4) at growth stage V2–V3 were transplanted into pots containing approximately 4.5 liters of SB-300 potting mix. At 25 days post-planting, the root zones of plants were infested with 1000 WCR eggs per pot. Twenty one days after infestation, individual plants were scored using the 0 to 3 root node-injury scale developed by Oleson et al.34. Negative controls consisted of a transgenic PHR03 null isoline. Statistical calculations were performed using JMP (Version 12. SAS Institute Inc., Cary, NC). To assess the root protection from corn rootworm feeding provided by dvssj1 in transgenic plants, the data were analyzed by non-parametric one-way ANOVA (Kruskall-Wallis; P < 0.0001). Dunnett’s post hoc test was used to compare all treatments against the control treatment, NULL.
DsRNA and siRNA northern blot analyses
Total RNA was extracted using the mirVana™ miRNA Isolation kit (Life Technologies, Carlsbad, CA) from T1 transgenic maize plants at leaf stage V6–V7. Ten μg of total RNA was fractionated on a 1.5% denaturing formaldehyde gel. For siRNA northern blot analysis, 20 μg of total RNA was fractionated on a 15% Criterion™ TBE-Urea Gel (Bio-Rad, Hercules, CA). RNAs were blotted to a Hybond-N+ membrane (Amersham, Little Chalfont, United Kingdom). The blots were pre-hybridized in ExpressHyb™ hybridization solution (Clontech, Mountain View, CA) for 1 h and then hybridized in the same solution as the DNA probe overnight at 65 °C for dsRNA and 37 °C for siRNA, respectively. The autoradiographs were digitized by ImageQuant™ LSA4000 (Fujifilm, Tokyo, Japan). Densitometry analyses of northern blots was peformed using Phoretixs 1D software (Cleaver Scientific, Rugby, UK).
Quantitative real-time PCR (qRT-PCR) and in situ hybridization (ISH)
Total RNA was extracted using Trizol and DNase I was used to remove genomic DNA. cDNA was synthesized from total RNA using the Bioline Sensifast cDNA kit (Taunton, MA) according to the manufacturer’s instructions. The designs of primers and probe regions are listed in Supplementary Table 3. Dvssj1 gene expression was quantified from WCR larvae collected after 12 and 48 h of feeding on diet incorporated with 0.5 ng μl−1 of dvssj1 frag1 dsRNA. Gene expression was analyzed using two-step real time quantitative RT-PCR. The assay was run, with 3 replicates per sample, using a single plex set up with Bioline Sensifast Probe Lo Rox kit (Taunton, MA) and analyzed using the 2−ΔΔCt method based on relative expression of the dvssj1 gene and a reference gene dvrps10. Data from qRT-PCR assays were analyzed using JMP (Version 12. SAS Institute Inc., Cary, NC) and statistical differences were detected using one-way analysis of variance (ANOVA) followed by Dunnett’s post-test; P < 0.05 was considered statistically significant.
For ISH analyses, target probes, preamplifier, amplifier, and label probe were designed by Advanced Cell Diagnostics (Hayward, CA). For chromogenic detection using 3,3′-Diaminobenzidine (DAB), label probe was conjugated to horseradish peroxidase (HRP). WCR were reared on artificial diet until 3rd instar,and then moved into individual wells containing diet incorporated with a dvssj1 frag1dsRNA and control dsRNA gus at 50 ng μl−1. Insects were collected 48 h post-treatment, fixed in 10% neutral buffered formalin (4% formaldehyde) for 48 to 72 h and processed for paraffin embedding. Paraffin sections were cut 4 μm thick, collected on Superfrost Plus slides (Fisher Scientific), air-dried overnight, and baked for 1 h at 60 °C. Sections were processed for RNA in situ hybridization with the RNAScope Detection Kit (Chromogenic) according to the manufacturer’s standard protocol (Advanced Cell Diagnostics, Hayward, CA). Slide images were acquired using a Leica Aperio® AT2 digital scanner and captured at 40× magnification with resolution of 0.25 μm pixel−1.
Western blot analysis
Western blot analysis was performed on solubilized WCR gut extracts from 3rd instar. The dissected gut tissue proteins were homogenized in buffer (50 mM Na2HPO4-NaH2PO4, 50 mM NaCl, 5 mM EGTA, 5 mM EDTA, pH 7.5) containing 2% Triton X100, 2 complete protease inhibitor cocktail tablets EDTA-free (Roche, Basel, Switzerland), and 1mM PMSF. The proteins were electrophoretically transferred to a nitrocellulose membrane and detected with either anti-DVSSJ1 mouse Ab (1:2,500, Genscript, USA) or anti-DVSSJ2 hybridoma supernatant mouse Ab (1:50, Genscript, USA), followed by goat anti-mouse (GAM)-HRP conjugated secondary Ab (Bio-Rad, Hercules, CA) (1: 12,500).
Microscopy
WCR eggs were reared on the same diet used for primary screening. Double-stranded RNA of dvssj1 frag1 was labelled with Cy3 by IVT and Cy3 fluorescence was used to confirm WCR feeding at a final concentration of 100 ng μl−1. Untreated control neonates were prepared by adding an equivalent volume of dsRNA buffer solution containing Cy3 only (no RNA). WCR neonates were transferred singly into wells at 24 h post-hatch and were collected after 72 h incubation with dsRNA and prepared for electron microscopy as described in the Supplementary Methods and by Rizzo et al.57 who have reported techniques for examination of ultrastructure via backscattered electron imaging in a scanning electron microscope.
Additional Information
Accession codes: The RNAi active target sequences have been deposited in the GenBank of National Center for Biotechnology Information under the accession number KU562965 (dvssj1); KU562966 (dvssj2); KU756279 (dvprotb); KU756280 (dvpat3) and KU756281 (dvrps10).
How to cite this article: Hu, X. et al. Discovery of midgut genes for the RNA interference control of corn rootworm. Sci. Rep. 6, 30542; doi: 10.1038/srep30542 (2016).
References
Gray, M. E., Sappington, T. W., Miller, N. J., Moeser, J. & Bohn, M. O. Adaptation and invasiveness of western corn rootworm: intensifying research on a worsening pest. Annu. Rev. Entomol. 54, 303–321, 10.1146/annurev.ento.54.110807.090434 (2009).
Levine, E. & Oloumi-Sadeghi, H. Management of Diabroticite Rootworms in Corn. Annu. Rev. Entomol. 36, 229–255, 10.1146/annurev.en.36.010191.001305 (1991).
Vaughn, T. A method of controlling corn rootworm feeding using a Bacillus thuringiensis protein expressed in transgenic maize. Crop Sci. 45, 931–938 (2005).
Deitloff, J., Dunbar, M. W., Ingber, D. A., Hibbard, B. E. & Gassmann, A. J. Effects of refuges on the evolution of resistance to transgenic corn by the western corn rootworm, Diabrotica virgifera virgifera LeConte. Pest Management Science, 10.1002/ps.3988 (2015).
Gassmann, A. J., Petzold-Maxwell, J. L., Keweshan, R. S. & Dunbar, M. W. Field-evolved resistance to Bt maize by western corn rootworm. PLos One 6, e22629, 10.1371/journal.pone.0022629 (2011).
Andow, D. A. et al. Early Detection and Mitigation of Resistance to Bt Maize by Western Corn Rootworm (Coleoptera: Chrysomelidae). J. Econ. Entomol. 109, 1–12. 10.1093/jee/tov238 (2016).
Fire, A. et al. Potent and specific genetic interference by double-stranded RNA in caenorhabditis elegans. Nature 391, 806–811, 10.1038/35888 (1998).
Kurreck, J. RNA interference: from basic research to therapeutic applications. Angew. Chem. Int. Ed. Engl. 48, 1378–1398, 10.1002/anie.200802092 (2009).
Price, D. R. & Gatehouse, J. A. RNAi-mediated crop protection against insects. Trends Biotechnol. 26, 393–400, 10.1016/j.tibtech.2008.04.004 (2008).
Timmons, L., Court, D. L. & Fire, A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263, 103–112, 10.1016/S0378-1119(00)00579-5 (2001).
Huvenne, H. & Smagghe, G. Mechanisms of dsRNA uptake in insects and potential of RNAi for pest control: A review. J. Insect Physiol. 56, 227–235, 10.1016/j.jinsphys.2009.10.004 (2010).
Belles, X. Beyond Drosophila: RNAi In Vivo and Functional Genomics in Insects. Annu. Rev. Entomol. 55, 111–128, 10.1146/annurev-ento-112408-085301 (2010).
Baum, J. A. et al. Control of coleopteran insect pests through RNA interference. Nature Biotech. 25, 1322–1326, 10.1038/nbt1359 (2007).
Siomi, M. C., Sato, K., Pezic, D. & Aravin, A. A. PIWI-interacting small RNAs: the vanguard of genome defence. Nat. Rev. Mol. Cell Biol. 12, 246–258 (2011).
Rangasamy, M. & Siegfried, B. D. Validation of RNA interference in western corn rootworm Diabrotica virgifera virgifera LeConte (Coleoptera: Chrysomelidae) adults. Pest Management Science 68, 587–591 (2012).
Alves, A. P., Lorenzen, M. D., Beeman, R. W., Foster, J. E. & Siegfried, B. D. RNA interference as a method for target-site screening in the Western corn rootworm, Diabrotica virgifera virgifera. J. Insect Sci. (Online) 10, 162, 10.1673/031.010.14122 (2010).
Baum, J. A. & Roberts, J. K. Progress Towards RNAi-Mediated Insect Pest Management. Adv. Insect Physiol. 47, 249–295, 10.1016/B978-0-12-800197-4.00005-1 (2014).
Scott, J. G. et al. Towards the elements of successful insect RNAi. J. Insect Physiol. 59, 1212–1221, 10.1016/j.jinsphys.2013.08.014 (2013).
Li, H. et al. Long dsRNA but not siRNA initiates RNAi in western corn rootworm larvae and adults. J. Appl. Entomol. 139, 432–445, 10.1111/jen.12224 (2015).
Ramaseshadri, P. et al. Physiological and cellular responses caused by RNAi- mediated suppression of Snf7 orthologue in western corn rootworm (Diabrotica virgifera virgifera) larvae. PLos One 8, e54270, 10.1371/journal.pone.0054270 (2013).
Beyenbach, K. W. & Wieczorek, H. The V-type H+ ATPase: molecular structure and function, physiological roles and regulation. J. Exp. Biol. 209, 577–589, 10.1242/jeb.02014 (2006).
Kim, D. W. et al. Differential physiological roles of ESCRT complexes in Caenorhabditis elegans. Mol. Cells 31, 585–592, 10.1007/s10059-011-1045-z (2011).
Dow, J. A. T. Insect midgut function. Adv. Insect Physiol. 19, 187–328 (1986).
Shanbhag, S. & Tripathi, S. Epithelial ultrastructure and cellular mechanisms of acid and base transport in the Drosophila midgut. J. Exp. Biol. 212, 1731–1744, 10.1242/jeb.029306 (2009).
Furuse, M. & Tsukita, S. Claudins in occluding junctions of humans and flies. Trends Cell Biology 16, 181–188, 10.1016/j.tcb.2006.02.006 (2006).
Behr, M., Riedel, D. & Schuh, R. The Claudin-like Megatrachea Is Essential in Septate Junctions for the Epithelial Barrier Function in Drosophila. Developmental Cell 5, 611–620, 10.1016/S1534-5807(03)00275-2 (2003).
Lane, N., Dallai, R., Martinucci, G. & Burighel, P. Electron microscopic structure and evolution of epithelial junctions. Molecular Mechanisms of Epithelial Cell Junctions: From Development to Disease. S. Citi, editor. RG Landes Company, Austin, TX 23, 43 (1994).
Izumi, Y. & Furuse, M. Molecular organization and function of invertebrate occluding junctions. Seminars in Cell & Developmental Biology 36, 186–193, 10.1016/j.semcdb.2014.09.009 (2014).
Tepass, U. & Tanentzapf, G. Epithelial cell polarity and cell junctions in Drosophila. Annu. Rev. Genet. 35, 747–784, 10.1146/annurev.genet.35.102401.091415 (2001).
Yanagihashi, Y. et al. Snakeskin, a membrane protein associated with smooth septate junctions, is required for intestinal barrier function in Drosophila. J. Cell Sci. 125, 1980–1990, 10.1242/jcs.096800 (2012).
Izumi, Y., Yanagihashi, Y. & Furuse, M. A novel protein complex, Mesh-Ssk, is required for septate junction formation in the Drosophila midgut. J. Cell Sci. 125, 4923–4933, 10.1242/jcs.112243 (2012).
Ciccarelli, F. D., Doerks, T. & Bork, P. AMOP, a protein module alternatively spliced in cancer cells. Trends Biochem. Sci. 27, 113–115 (2002).
Chien, C. H., Chen, W. W., Wu, J. T. & Chang, T. C. Investigation of lipid homeostasis in living Drosophila by coherent anti-Stokes Raman scattering microscopy. J. Biomed. Opt. 17, 126001, 10.1117/1.jbo.17.12.126001 (2012).
Oleson, J. D., Park, Y. L., Nowatzki, T. M. & Tollefson, J. J. Node-Injury Scale to Evaluate Root Injury by Corn Rootworms (Coleoptera: Chrysomelidae). J. Econ. Entomol. 98, 1–8 (2005).
Zhao, L.-N. et al. Elongation Factor 1β′ Gene from Spodoptera exigua: Characterization and Function Identification through RNA Interference. International J. Molecular Sci. 13, 8126–8141, 10.3390/ijms13078126 (2012).
Li, X., Zhang, M. & Zhang, H. RNA Interference of Four Genes in Adult Bactrocera dorsalis by Feeding Their dsRNAs. PLos ONE 6, e17788, 10.1371/journal.pone.0017788 (2011).
Broglie, K. E., Kriss, K., Lu, A. L., Muthalagi, M. & Presnail, J. K. Inventors; E.I. DuPont De Nemours And Company, assignee. Compositions and methods to control insect pests. United States patent US20110054007 A1 (3 Mar 2011).
Hu, X. et al. Inventors; E.I. DuPont De Nemours And Company, assignee. Compositions and methods to control insect pests. United States patent US20140275208 A1 (18 Sep 2014).
Attrill, H. et al. FlyBase: establishing a Gene Group resource for Drosophila melanogaster. Nucleic Acids Res 44, D786–792, 10.1093/nar/gkv1046 (2016).
Gutierrez, E., Wiggins, D., Fielding, B. & Gould, A. P. Specialized hepatocyte-like cells regulate Drosophila lipid metabolism. Nature 445, 275–280, 10.1038/nature05382 (2007).
Endo, Y. & Nishiitsutsuji-Uwo, J. Fine structure of development endocrine cells and columnar cells in the cockroach midgut. Biomed. Res. 3, 637–644 (1982).
Hakim, R. S., Baldwin, K. & Smagghe, G. Regulation of midgut growth, development, and metamorphosis. Annu. Rev. Entomol. 55, 593–608, 10.1146/annurev-ento-112408-085450 (2010).
Spence, J. R., Lauf, R. & Shroyer, N. F. Vertebrate intestinal endoderm development. Developmental Dynamics 240, 501–520, 10.1002/dvdy.22540 (2011).
Dun, Z., Mitchell, P. D. & Agosti, M. Estimating Diabrotica virgifera virgifera damage functions with field trial data: applying an unbalanced nested error component model. J. Appl. Entomol. 134, 409–419, 10.1111/j.1439-0418.2009.01487.x (2010).
Tinsley, N. A., Estes, R. E. & Gray, M. E. Validation of a nested error component model to estimate damage caused by corn rootworm larvae. J. Appl. Entomol. 137, 161–169, 10.1111/j.1439-0418.2012.01736.x (2013).
Tinsley, N. A. et al. Estimation of efficacy functions for products used to manage corn rootworm larval injury. J. Appl. Entomol. 140, 414–425, 10.1111/jen.12276 (2016).
Hong, B., Nowatzki, T. M., Sult, T. S., Owens, E. D. & Pilcher, C. D. Sequential sampling plan for assessing corn rootworm (Coleoptera: Chrysomelidae) larval injury to Bt maize. Crop Protection 82, 36–44, 10.1016/j.cropro.2016.01.001 (2016).
Thompson, J. D., Higgins, D. G. & Gibson, T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 (1994).
Finn, R. D. et al. Pfam: the protein families database. Nucleic Acids Res. 42, D222–230, 10.1093/nar/gkt1223 (2014).
Hirokawa, T., Boon-Chieng, S. & Mitaku, S. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14, 378–379 (1998).
Zhao, J.-Z. et al. mCry3A-selected western corn rootworm (Coleoptera: Chrysomelidae) colony exhibits high resistance and has reduced binding of mCry3A to midgut tissue. J. Econ. Entomol. 109, 1369–1377, 10.1093/jee/tow049 (2016).
Finney, D. J. Probit Analysis, 3rd ed. Cambridge University Press, New York (1971).
Sambrook, J. & Russell, D. W. Molecular cloning a laboratory manual (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, 2001).
Callis, J., Fromm, M. & Walbot, V. Introns increase gene expression in cultured maize cells. Genes Dev. 1, 1183–1200 (1987).
Cho, M. J. et al. Agrobacterium-mediated high-frequency transformation of an elite commercial maize (Zea mays L.) inbred line. Plant Cell Rep. 33, 1767–1777, 10.1007/s00299-014-1656-x (2014).
Zhi, L. et al. Effect of Agrobacterium strain and plasmid copy number on transformation frequency, event quality and usable event quality in an elite maize cultivar. Plant Cell Rep. 34, 745–754, 10.1007/s00299-014-1734-0 (2015).
Rizzo, N. W., Duncan, K. E., Bourett, T. M. & Howard, R. J. Backscattered electron SEM imaging of resin sections from plant specimens: observation of histological to subcellular structure and CLEM. J. Microscopy (Published Online), 10.1111/jmi.12373 (2015).
Acknowledgements
We thank Adane Kassa and Jeff Robson for bioassay and vector construction support; DuPont Pioneer maize transformation and controlled environment groups for generating and managing transgenic plants; Zaiqi Pan for statistical analysis; Deirdre Kapka-Kitzman for providing WCR midgut homogenates; Brooke Peterson-Burch for bioinformatics support; John Ding for protein expression; Todd Hauser and Paul Oliver for ISH support; Ian Lamb and Timothy Nowatzki for thoughtful comments and suggestions.
Author information
Authors and Affiliations
Contributions
X.H., J.Z.Z., R.J.H. and J.K.P. conceived and designed the study. N.M.R., K.E.D., X.P., L.A.P., M.A.O., B.M.K., J.P.S., V.C.C., G.S. and J.L.R. performed the data analysis and interpreted the results. X.H., A.L.L. and G.W. with input from all of the authors wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Hu, X., Richtman, N., Zhao, JZ. et al. Discovery of midgut genes for the RNA interference control of corn rootworm. Sci Rep 6, 30542 (2016). https://doi.org/10.1038/srep30542
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep30542
This article is cited by
-
Basal stem cell progeny establish their apical surface in a junctional niche during turnover of an adult barrier epithelium
Nature Cell Biology (2023)
-
Designing Climate-Resilient Crops for Sustainable Agriculture: A Silent Approach
Journal of Plant Growth Regulation (2023)
-
Characterization of DvSSJ1 transcripts targeting the smooth septate junction (SSJ) of western corn rootworm (Diabrotica virgifera virgifera)
Scientific Reports (2020)
-
RNAi for management of Asian long-horned beetle, Anoplophora glabripennis: identification of target genes
Journal of Pest Science (2020)
-
Development of CS-TPP-dsRNA nanoparticles to enhance RNAi efficiency in the yellow fever mosquito, Aedes aegypti
Scientific Reports (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.